October 7, 2018
A new tool developed by Weizmann Institute of Science researchers for mapping cells reveals how complex regulatory systems evolved to help diverse cell types cooperate.
The first multi-celled organisms began appearing on Earth hundreds of millions of years ago. That is, once-independent cells gradually traded autonomy for a collective lifestyle.
To understand this process, the researchers tracked down some of the simplest animals on the ‘tree of life’ and inspected the cell types contained in each animal as well as the regulatory mechanisms that these cells use to cooperate.
As reported in two recent papers, Professsor Amos Tanay and his group achieved new insight into this moment in evolutionary time with some cutting-edge methods they developed for mapping the functions of thousands of individual cells.
Dr Arnau Sebe-Pedros, a postdoctoral fellow in Tanay’s group in the Computer Science and Applied Mathematics, and Biological Regulation Departments at the Institute, decided to focus on four kinds of little-known marine animals: cnidarians, ctenophores, sponges and placozoans. These animal lineages, at the base of our common animal tree of life, contain important clues about the earliest steps of animal evolution.
Although many others have attempted to elucidate this particular moment in evolution, Sebe-Pedros had at his disposal a unique tool: A way of mapping each cell in an organism down to its molecular makeup.
“This tool had already been used to map cells in such model animals as mice and fruit flies. Even in these well-studied animals it yielded some surprises,” said Tanay.
Sebe-Pedros added: “There are huge areas of the ‘tree of life’ that we have yet to explore.”
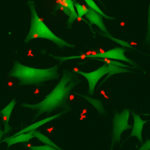
Tanay, Sebe-Pedros and the team began with a relatively familiar organism – a sea anemone called Nematostella vectensis. This genome of this species had been sequenced in 2007, and the surprise was how similar it was to that of humans, including hundreds of shared genes. Although sea anemones are not your typical lab animal, there are a number of research groups using them as a model for studies of development and evolution. Tanay and his group joined forces with one of these experts – Dr Heather Marlow of the Pasteur Institute in Paris – and the two groups collaborated on a study of these organisms, which was reported in Cell. The researchers found that these animals have an unusually sophisticated genomic and cellular makeup.
This slender body, topped by a crown of clear, stinging tentacles, contains dozens of different cell types; of these, some 30 are different neuronal cell types that form a relatively complex nervous system.
Emboldened by their success with the sea anemone, the group turned to creatures even more distant from us on the evolutionary tree and less well-understood.
Sebe-Pedros contacted colleagues around the world in a request for species, ultimately finding several that were definitively not lab animals: Not only could some these not be grown in the lab, they could only be collected in certain places, at certain times of the year.
But the Weizmann team and their collaborators managed to assemble a variety of unusual samples, among them a species of sponge from the Australian Great Barrier Reef; a comb jelly also known as a sea walnut, which lives on the northern coasts of the Atlantic Ocean and the Mediterranean Sea; and the placozoan Trichoplax, an animal first discovered in the reefs near Eilat in the 1970s that looks like a millimeter-sized pancake. Like sea anemones, these organisms are often considered to be simple, in part because they never developed the bilateral symmetry of most living animals.
As the team reported in Nature Ecology and Evolution, the complexity of the genome’s regulatory mechanisms and the number of cell types in these species seems to go hand-in-hand. The fewer types of cell each had, the more the genes within each cell were regulated in a way that resembles that of single-celled organisms. The chains of events for turning various genes on or off was more direct in the simpler creatures and more sophisticated in those with many cell types. Thus placozoans, with only a few cell types, had very simple systems of gene regulation; while the sea anemones exhibited genomic regulation closer to that of the more complex animals – hinting at how complex regulatory systems co-evolved with diversification into a broader range of cell types.
The researchers uncovered quite a few previously unknown cell types in each of the animals examined. Even in placozoans, which do not have neurons, the study revealed a number of specialized cell types that produce regulatory peptides. In the absence of a nervous system, these unique peptides coordinate elaborate, collective cell behaviours.
“We created a tool that enables us to choose the less-studied organisms and to get an accurate picture of how the genes are activated in each cell. This massive increase in our ability to samples diverse organisms provides a new basis for comparison, opening up the branches of the evolutionary tree of life to exciting new discoveries,” concluded Tanay.
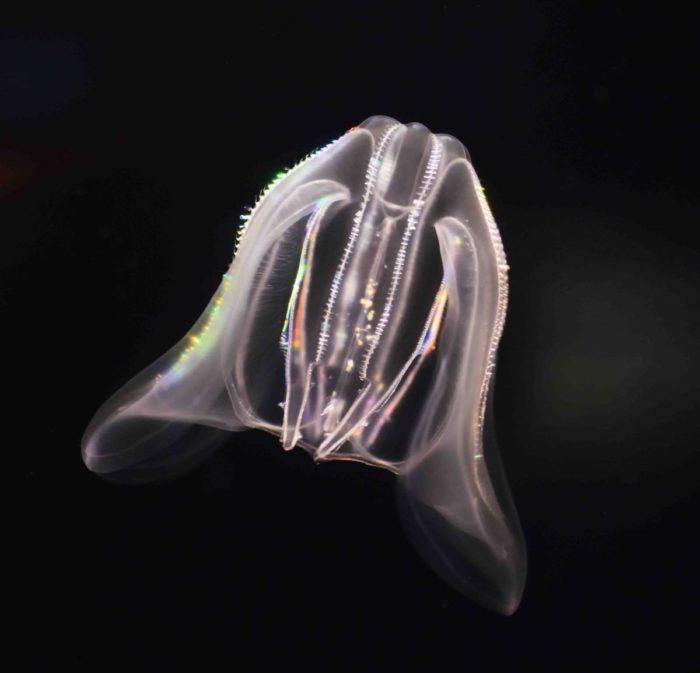
The "sea walnut" comb jelly
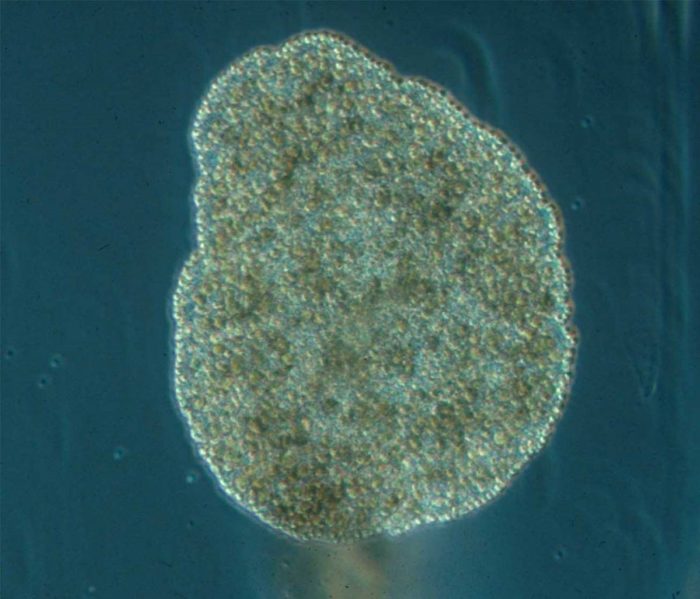
The pancake-shaped placozoan has peptides that coordinate complex collective cell behaviours
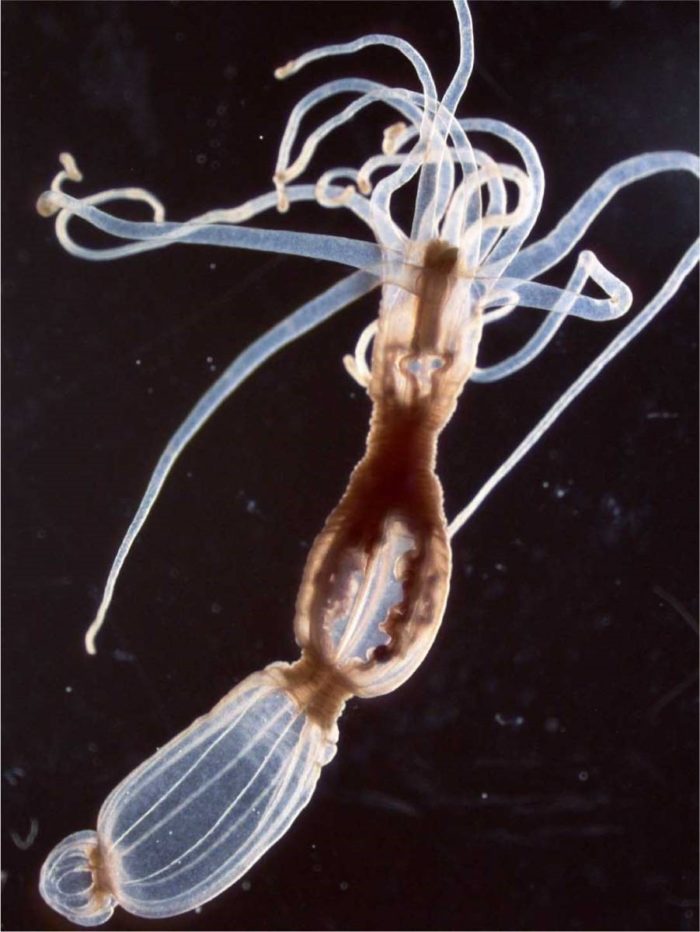
The Nematostella sea anemone has a relatively complex nervous system

(l-r) Dr Elad Chomsky, Dr Zohar Mukamel, Dr Arnau Sebe-Pedros and Professor Amos Tanay