
October 7, 2016
Discovering how proteins originated and developed is the focus of research in the lab of Professor Dan Tawfik of the Weizmann Institute’s Biomolecular Sciences Department, and where, with the help of computer science, they went back in time to find the answers.
Made up of hundreds of amino acids lined up in the correct sequence and folded into a well-defined shape to enable function, proteins are complex structures, so how they are assembled was a key question.
Professor Tarik’s team investigated a class of proteins known as propellers, which as their name suggests, may have originally been assembled from the welding together of a number of ‘blades’.
The proteins we know today are all the result of natural selection: mistakes in DNA, or mutations, in the genes occasionally making useful changes in the protein’s structure or function.
But before this evolutionary tinkering could take place, the ancestors of those proteins had to make a leap to get from a bunch of chemical compounds – amino acids, or short peptides (tethered amino acids) – to functional, living molecules. Peptides have been known to stick to one another; but could self-assembly, as some have suggested, lead to a whole, functional protein?
Tawfik compares the idea of the self-assembly of a modern protein from amino acids to an airplane assembling itself from the parts lying around a hangar. He hypothesizes that there was an intermediate step.
“Peptides are relatively short amino acid sequences that have defined structures; and these structures have been known to arise spontaneously both in nature and in the lab. We asked whether, under certain circumstances, peptides could fuse together, the same forces that hold the peptides in their folded form acting to bind the pieces together and even enabling them to function,” he says.
For their investigation, the researchers chose a family of proteins in which repeating sub-units look like the blades of a propeller, these all joined in the middle. They then went ‘back in time’ to retrace the evolution of this protein.
For this task, they used a computational approach that had been pioneered by Linus Pauling to compute, for each sequence, the one that had preceded it. This method eventually led them to a single peptide ‘blade’ – a sequence only 50 amino acids in length; several of these could self-assemble to form a larger structure. The researchers hypothesised that this structure may have been the primordial ancestor of today’s propeller protein.
To then observe the ‘time machine’ in reverse – that is, to see how a protein could have evolved from peptides – the research group turned to the lab.
With the directed evolution techniques in Tawfik’s lab, sequences undergo a speeded-up version of the natural mutation-and-natural-selection process. After the 50-amino-acid sequences underwent a few generations of directed evolution, they gained the ability to connect to one another, eventually producing modern-looking proteins made up of 250 amino acids. Since they were able to track this process step by step – backwards and forwards – the researchers were able to pinpoint the forces that shape the process of protein genesis.
Did propeller proteins actually come into being this way? The researchers looked at bacteria to see if evidence for this sort of evolution could be reconstructed from their genomes. Now that they knew the ‘parts’ of the propeller protein and how they could have developed, the researchers were able to identify sequences corresponding to modern propeller proteins, in which the process of duplication and fusion of blades was evident, as well as a part of another protein from which this blade was ‘co-opted’ to give birth to a new propeller.
Dr Paola Laurino and Professor Dan Tawfik investigate proten evolution backwards and forwards
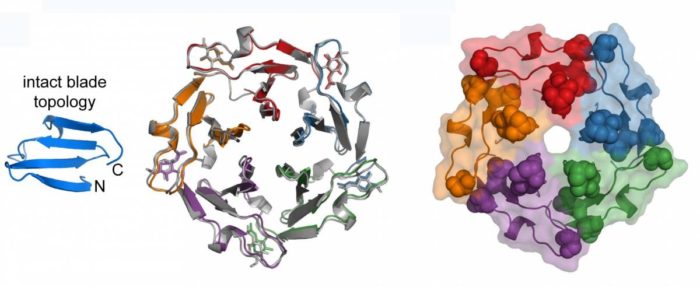
Blades stick together: the proposed evolution of a complete protein






