July 24, 2019
Researchers at the Weizmann Institute of Science have shown in mice that intestinal microbes, collectively termed the gut microbiome, may affect the course of amyotrophic lateral sclerosis (ALS), also known as Lou Gehrig’s disease, which damages motor neurons in the brain and spinal cord.
As reported recently in Nature, progression of an ALS-like disease was slowed after the mice received certain strains of gut microbes or substances known to be secreted by these microbes. Preliminary results suggest that the findings on the regulatory function of the microbiome may be applicable to human patients with ALS.
“Our long-standing scientific and medical goal is to elucidate the impact of the microbiome on human health and disease, with the brain being a fascinating new frontier,” said Professor Eran Elinav of Weizmann’s Immunology Department.
His team performed the study together with that of Professor Eran Segal of the Computer Science and Applied Mathematics Department.
“Given increasing evidence that microbiome affects brain function and disease, we wanted to study its potential role in ALS,” said Segal.
The study was led by postdoctoral fellows Drs Eran Blacher and Stavros Bashiardes, and by staff scientist Dr Hagit Shapiro, all in the Elinav lab. They collaborated with Dr Daphna Rothschild, a postdoctoral fellow in Eran Segal’s lab, and Dr Marc Gotkine, Head of the Motor Neuron Disease Clinic at the Hadassah Medical Center, as well as with other scientists from Weizmann and elsewhere.
The scientists started out demonstrating in a series of experiments that the symptoms of an ALS-like disease in transgenic mice worsened after these mice were given broad-spectrum antibiotics to wipe out a substantial portion of their microbiome. In addition, they found that growing these ALS-prone mice in germ-free conditions (in which, by definition, mice carry no microbiome of their own), is exceedingly difficult, as these mice had a hard time surviving in the sterile environment. Together, these results hinted at a potential link between alterations in the microbiome and accelerated disease progression in mice that were genetically susceptible to ALS.
Next, using advanced computational methods, the scientists characterised the composition and function of the microbiome in the ALS-prone mice, comparing them to regular mice. They identified 11 microbial strains that became altered in ALS-prone mice as the disease progressed or even before the mice developed overt ALS symptoms. When the scientists isolated these microbial strains and gave them one by one – in the form of probiotic-like supplements – to ALS-prone mice following antibiotic treatment, some of these strains had a clear negative impact on the ALS-like disease. But one strain, Akkermansia muciniphila, significantly slowed disease progression in the mice and prolonged their survival.
To reveal the mechanism by which Akkermansia may be producing its effect, the scientists examined thousands of small molecules secreted by the gut microbes. They zeroed in on one molecule called nicotinamide (NAM): Its levels in the blood and in the cerebrospinal fluid of ALS-prone mice were reduced following antibiotic treatment and increased after these mice were supplemented with Akkermansia, which was able to secrete this molecule. To confirm that NAM was indeed a microbiome-secreted molecule that could hinder the course of ALS, the scientists continuously infused the ALS-prone mice with NAM. The clinical condition of these mice improved significantly. A detailed study of gene expression in their brains suggested that NAM improved the functioning of their motor neurons.
Finally, the team examined the microbiome and metabolite profiles of 37 human ALS patients and compared them to those of family members sharing the same household. A detailed genomic analysis suggested that the gut microbiomes of ALS patients were distinct in composition and functional features from those of healthy controls. In particular, numerous microbial genes involved in the synthesis of NAM were significantly suppressed in ALS patients.
An analysis of thousands of small molecules in the blood also revealed a distinct pattern in ALS patients as compared to controls. Here too, many of the intermediary molecules involved in the NAM synthesis were altered in the blood of ALS patients. When the researchers tested the levels of NAM itself, they found these to be significantly reduced in both the blood and the brain of 60 human ALS patients as compared to controls. Moreover, there was a correlation between reduced NAM levels and the degree of muscle weakness in the patients.
“These findings are only a first step towards achieving a comprehensive understanding of the potential impact of the microbiome on ALS, but they suggest that in the future, various means of altering the microbiome may be harnessed for developing new therapeutic options for ALS,” said Elinav,
Also taking part in the study were Uria Mor, Dr Mally Dori-Bachash, Dr Christian Kleimeyer, Claudia Moresi, Yotam Harnik, Maya Zur, Rotem Ben-Zeev Brik, Dr Denise Kviatcovsky, Dr Niv Zmora, Yotam Cohen and Dr Nira Amar of the Immunology Department; Noam Bar and Izhak Levi of the Molecular Cell Biology Department and Computer Science and Applied Mathematics Department; Professor Michal Schwartz of the Neurobiology Department; Tevie Mehlman and Dr Alexander Brandis of the Life Sciences Core Facilities Department; Dr Inbal Biton, Dr Yael Kuperman, Dr Michael Tsoory and Professor Alon Harmelin of the Veterinary Resources Department; Michal Zabari of the Department of Neurology, Hadassah-Hebrew University Medical Center; Leenor Alfahel and Professor Adrian Israelson of the Department of Physiology and Cell Biology, Ben-Gurion University of the Negev; and Dr Liisa Arike, Dr Malin E.V. Johansson and Professor Gunnar C. Hansson of the Department of Medical Biochemistry and Cell Biology, University of Gothenburg, Sweden.
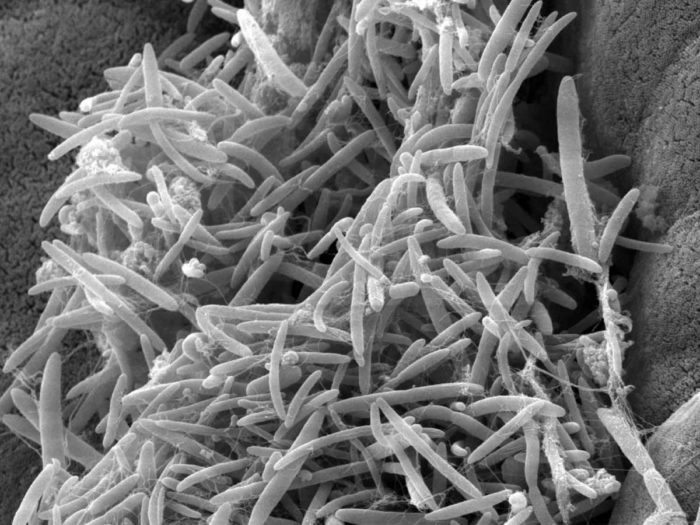
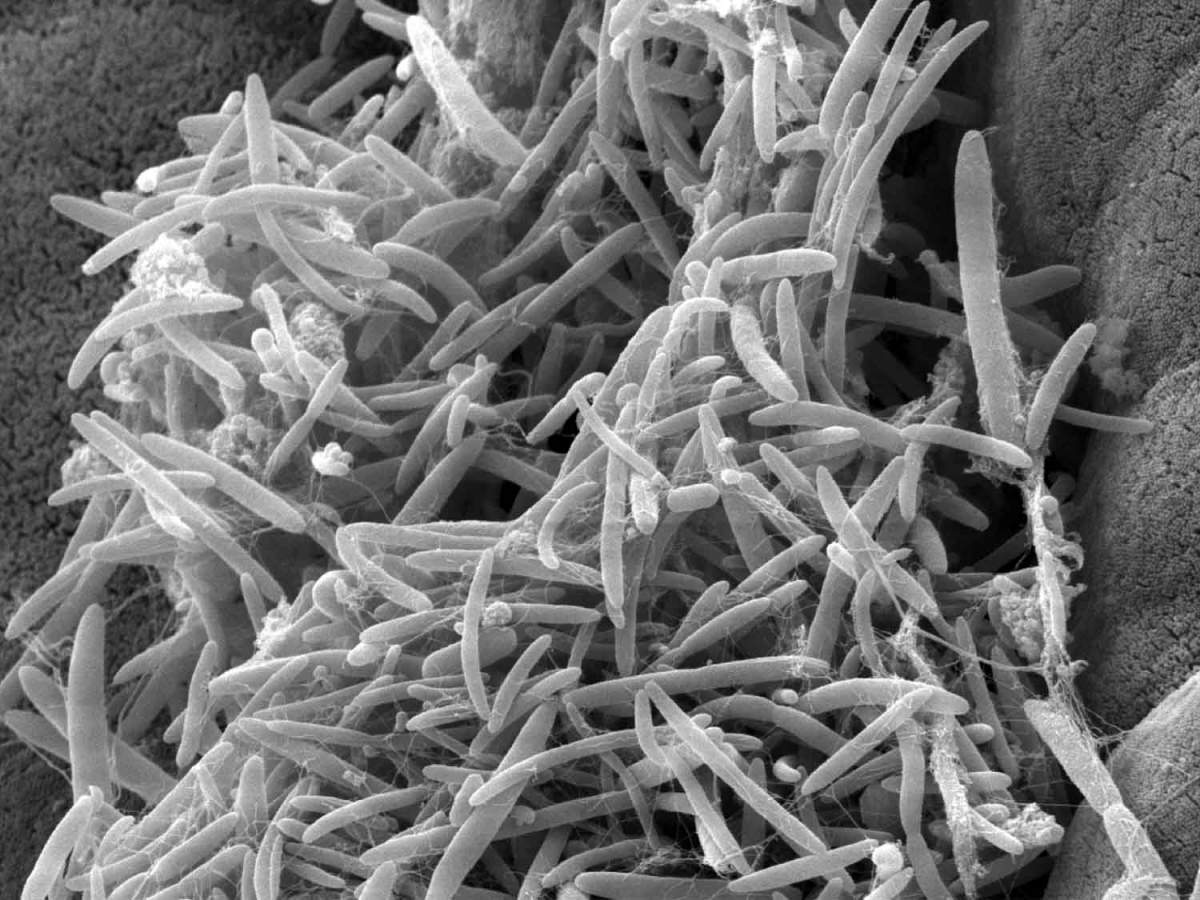
Gut microbes such as these were found to have altered levels in ALS patients
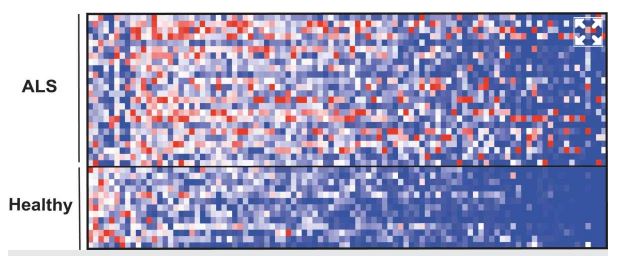
The repertoire of small molecules (metabolites) in blood, many of which originate in the microbiome, showed a different pattern in patients with ALS (top) compared with healthy individuals (bottom)