April 25, 2020
Scientists at the Weizmann Institute of Science have produced a cellular factory-on-a-chip which could be used to design, produce and test drugs against antibiotic resistant bacteria.
The ribosome is a cell’s protein factory and is the only natural machine that manufactures its own parts. That is why understanding how the machine is made could unlock the door to everything from understanding how life develops to designing new methods of drug production.
A seven year research effort at the Weizmann Institute of Science has resulted in demonstrating the self-synthesis and assembly of the small subunit of a ribosome (30S) on the surface of a chip, recently published in Science Advances.
Led by Professor Roy Bar-Ziv and Staff Scientist Dr Shirley Shulman Daube of the Institute’s Chemical and Biological Physics Department, one of the main challenges to their project was the sheer number of different molecules the cell must produce to make the subunit: The core is a long strand of RNA, and 20 different proteins must be attached to the strand.
These get organised by the weak chemical forces between the protein molecules and the RNA – repelling at some points and attracting in others – and the whole structure then relies on the proper manufacture and organisation of each component.
Add to that another six proteins that are not part of the structure, but act as chaperones to assist in the assembly. That makes at total of at least 27 different genes – one to encode each component or chaperone – that must work together to make the subunit.
Together with postdoctoral fellow Dr Michael Levy, who led the current study, and research student Reuven Falkovich, the team produced the subunits on tailored chips that Bar-Ziv developed in his lab. Ultimately, they succeeded in mimicking the natural process of synthesizing the parts and assembling them into the ribosome subunits. The tiny chips in Bar-Ziv’s lab are based on densely-packed DNA strands attached at one end to the surface.
In the beginning, the team used all 27 genes needed to reproduce the 30S subunit of a ribosome from an E coli bacterium. The components were caught in “molecular traps” placed near their genes, and this improved the efficiency of the process and enabled the scientists to observe the production process in real time. Then they took a step back, allowing the various parts to autonomously assemble themselves into the ribosomal units, without outside direction or interference.
Molecular hierarchy
In the beginning, Bar-Ziv and Shulman Daube found they could make the components, but getting them to self-assemble, as the natural structures do, was a challenging hurdle. Over the course of the next seven years and hundreds of trials, the scientists tracked down the proper placement of the genes on the chips. Something like the organization of genes in the chromosome, the genes on the chip had to be positioned in the right locations, and in the proper relative quantities.
This, it turned out, was crucial to the overall orchestration of the complex assembly process. Each time, the scientists would attach a different constellation of genes to the chips, narrowing down the possibilities until they had a composition that could mimic that natural process of subunit production as well as self-assembly. In nature, subunit assembly is a hierarchal process. In the course of their experiments, the scientists were able to break down the assembly to the individual steps to prove that the end result was a self-assembled subunit, and to observe the roles of the chaperones in this process.
Bar-Ziv and Shulman Daube believe that this new insight into producing complex, multi-component structures could pave the way to creating all sorts of other complex, molecular structures – existing ones as well as those not yet found in nature. Thus, for example, structures found in disease-causing bacteria might be produced for the purpose of safely testing and manufacturing drugs, vaccines or diagnostics without using whole, infectious bacteria. In the future, the self-assembly method could lead to the development of new kinds of vaccines, as well as assembly lines for various complex molecules for different industries.
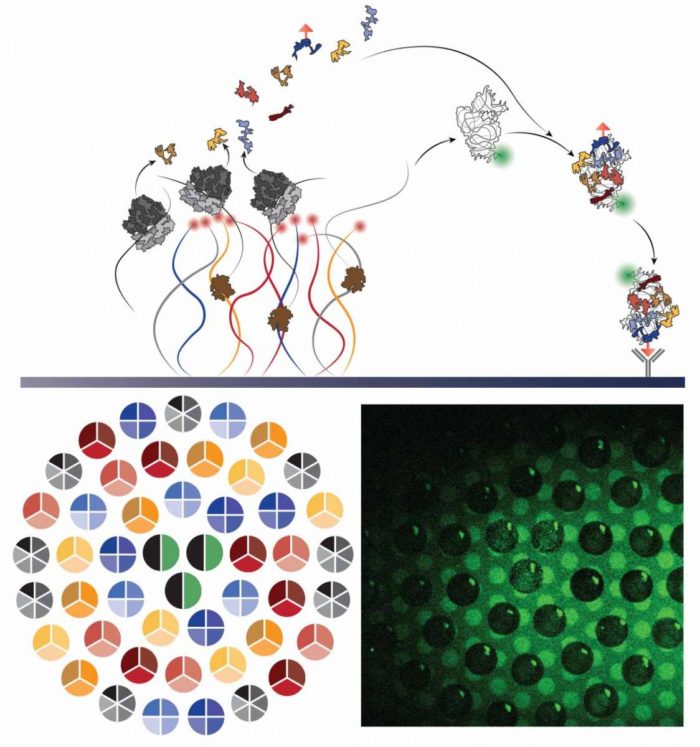
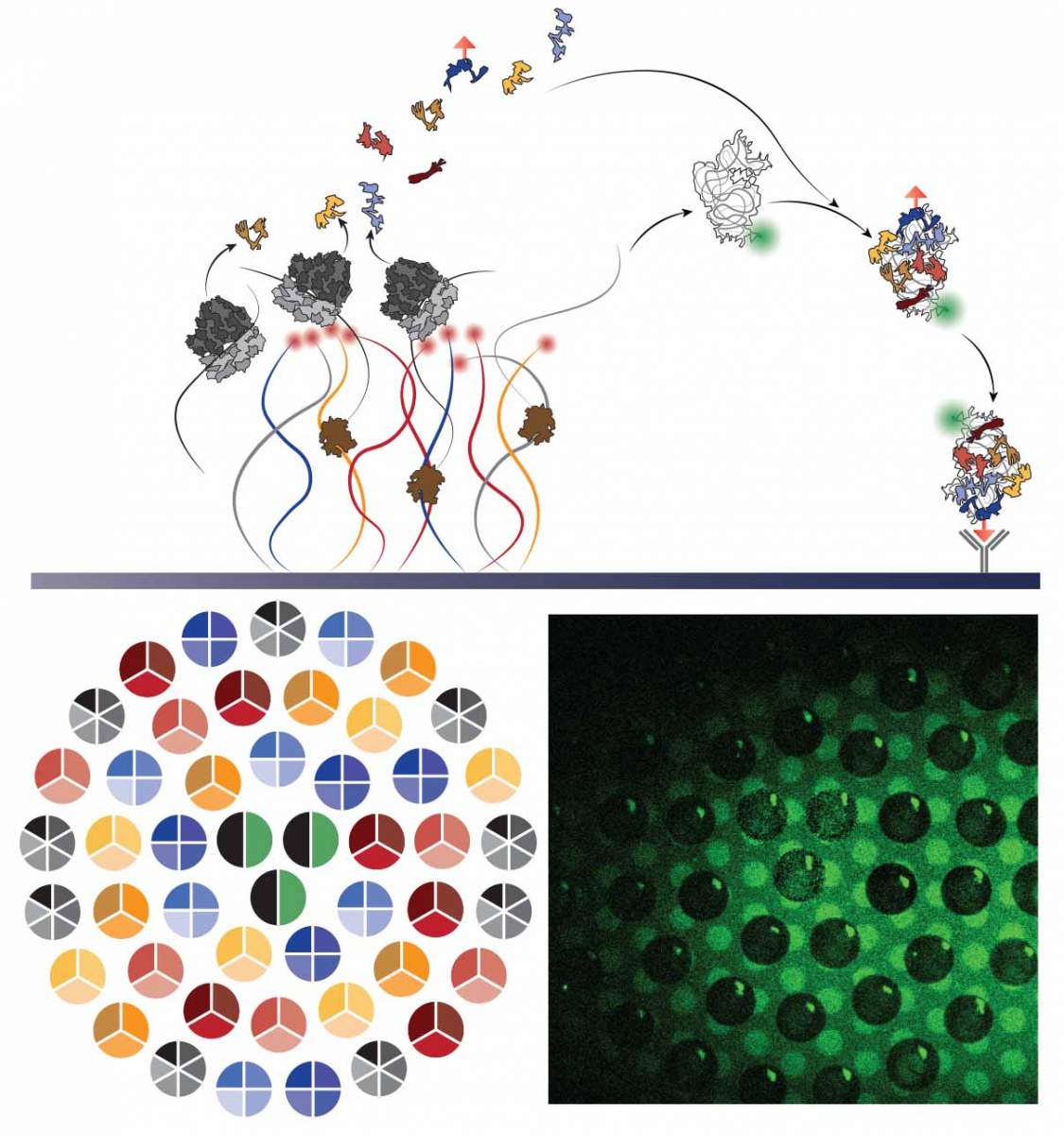
Visual representation of the self-assembly process: Synthesis of proteins and ribosomal RNA from synthetic DNA strands attached to a chip leads to the self-assembly of a new ribosomal subunit on the surface of the chip. Bottom left: Imaging of DNA strands grouped into several densely-packed "brushes" in the shape of circles; right: fluorescence imaging of a "carpet" of subunits at the completion of the assembly process
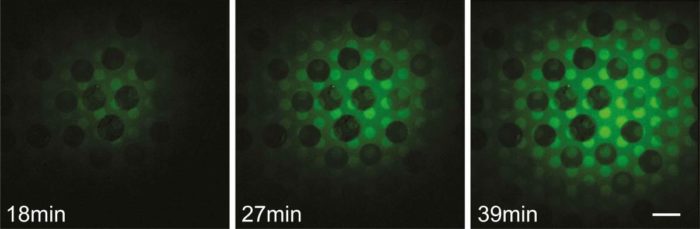
Fluorescence imaging of the self-assembly process, which takes about 40 minutes to complete





